Lipidomics on nanoneedles
We assessed the suitability of nanoneedles for DESI-MSI lipidomics, using nanoneedle arrays with 2 μm pitch, 4.00 μm height, 50 nm tip diameter, 600 nm base diameter, 77.2 m2 g−1 surface area, 50% porosity and 7.9 nm average pore diameter, on 8 × 8 mm silicon substrates (Supplementary Fig. 1). The analysis of porcine brain lipid extract revealed that nanoneedles and flat surfaces offer comparable sensitivity for the most representative lipids, consistent with established DESI limits of detection (LODs)19 (Extended Data Fig. 1a,b and Supplementary Table 1). Nanoneedle substrates typically displayed slightly higher LODs than flat silicon, suggesting that nanotextured surfaces can dampen the desorption–ionization process. However, the relative peak intensity correlated well between nanoneedles and flat substrates, indicating that the analysis on nanoneedles accurately recapitulated the lipid composition of the mixture (Extended Data Fig. 1c and Supplementary Fig. 2). Optimization of DESI-MSI analytical parameters20 for nanoneedles significantly increased the number of detectable features by a factor of 2.47, from 408 ± 83 to 1,007 ± 141 (Extended Data Fig. 1d–f).
Lipidomics imaging of molecular replicas
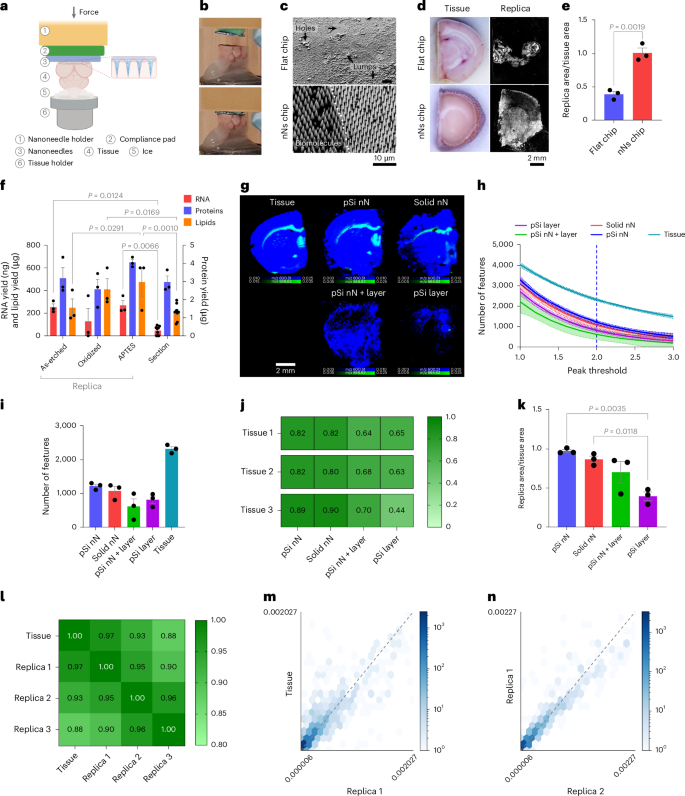
We then established the ability of nanoneedles to generate a molecular replica by imprinting frozen mouse brain (Fig. 2a,b). Computational modelling indicated that, during imprinting, a few micrometres of tissue transiently thaw, allowing collection of biomolecules (Supplementary Fig. 3). The resulting replica formed a uniform molecular adsorbate on the nanoneedles, whereas using a flat substrate yielded poor adsorbate uniformity, presenting areas without coverage and areas of adsorbate accumulation (Fig. 2c). The nanoneedle replica fully reproduced the original tissue and macroscopically retained its morphology, whereas the flat replica reproduced only less than half of the tissue, lacking clear morphology (Fig. 2d,e).
a, Schematics of the nanoneedle imprinting system. Created with BioRender.com. b, Photographs illustrating the system approaching (top) and imprinting (bottom) nanoneedles on tissue. c, Scanning electron microscopy images of the molecular adsorbate on flat and nanoneedle substrates after imprinting. d, Photographs of the imprinted tissue and fluorescence microscopy images of the corresponding molecular replica. e, Quantification of replica-to-tissue area ratio. Data are mean ± s.e.m. (N = 3 independent biological replicates). Statistics: two-tailed unpaired t-test. f, Quantification of RNA, lipids and proteins from nanoneedle replicas with different surface chemistries (as-etched, APTES, oxidized) and a 10-µm brain tissue section. ata are mean ± s.e.m. (N = 3 independent biological replicates). Statistics: two-way ANOVA with a post-hoc Tukey’s multiple-comparisons test. g, DESI-MSI maps of grey matter (m/z 834.53, blue) and white matter (m/z 888.62, green) lipids from a tissue slice and molecular replicas generated using porous silicon nanoneedles (pSi nN), solid silicon nanoneedles (solid nN), porous silicon nanoneedles over a porous silicon layer (pSi nN + layer) and a porous silicon layer (pSi layer). h, Quantification of the number of features as a function of the confidence threshold set for peak identification in samples from g. Data are mean ± s.e.m. (N = 3 independent biological replicates). i, The feature count at a peak threshold of 2.0 (95% confidence) for samples from g. Data are mean ± s.e.m. (N = 3 independent biological replicates). j, The correlation matrix of spectra from three tissue sections and their corresponding replicas from g. k, The relative surface area of molecular replicas to original tissue for samples from g. Data are mean ± s.e.m. (N = 3 independent biological replicates). Statistics: ordinary one-way ANOVA with post-hoc Tukey’s multiple-comparisons test. l, A correlation matrix comparing three pSi nN replicas with the original tissue section. m, A correlation scatter plot of lipids peak intensities between the tissue section and the first replica. n, A correlation scatter plot of lipid peak intensities between the first and second replica.
We then evaluated the composition of the molecular adsorbate on replicas generated from nanoneedles as-etched, O2-plasma oxidized and (3-aminopropyl)-triethoxysilane (APTES) functionalized, assessing the role of surface polarity and charge on biomolecular collection efficiency21,22. Oxidation and APTES respectively generated negative and positive charges, providing more polar surfaces than the as-etched porous silicon. For all the functionalizations considered, the amount of collected RNA, proteins and lipids compared favourably with those that could be extracted from the reference tissue (Fig. 2f). RNA collection ranged between 210 ng and 350 ng, while protein collection ranged between 2.5 μg and 4.3 μg. Lipid collection was more efficient on APTES and oxidized nanoneedles compared with as-etched ones, ranging between 200 μg and 600 μg. These data indicated that plasma oxidized nanoneedles were suitable for spatial lipidomics, because their lipid collection efficiency of 410 ± 95 μg was well within the range in which a large number of features were detectable (Fig. 2f) and compared favourably with a 10-μm tissue section (210 ± 23 μg), known to be imageable by DESI-MSI. The use of oxidized nanoneedles also mitigated potential concerns regarding interference from the desorption of polymerized APTES.
We also compared the quality of the replica generated using porous silicon nanoneedles (pSi nN) with those obtained using solid silicon nanoneedles (solid nN), porous silicon nanoneedles over a continuous porous silicon layer (pSi nN + layer) and a continuous porous silicon layer (pSi layer)23,24,25 (Supplementary Fig. 4). pSi nN displayed the largest number of features and the least variability, followed by the solid nN, while the pSi nN + layer and pSi layer yielded less features with higher variability (Fig. 2g–i). The pSi nN also outperformed the other substrates in their spectral correlation to the original tissue, closely matched by solid nN (Fig. 2j), and most closely reproduced the surface area of the original tissue (Fig. 2g,k).
The imprinting was reproducible, with three replicas from the same surface preserving morphological features and showing strong correlation with the reference section and with each other across the whole range from low- to high-abundance lipids (Fig. 2l–n and Supplementary Fig. 5).
These data indicated that a molecular replica generated using porous silicon nanoneedles could accurately and robustly reproduce the lipidomic profile of a tissue. However, the presence of a continuous porous layer reduced the efficiency of the desorption–ionization process, favouring the use of substrates with nanoneedles only. In addition, porous nanoneedles marginally outperformed solid ones, producing replicas that more closely represented the original tissue in terms of area, number of features and correlation.
Spatial lipidomics of murine glioma replicas
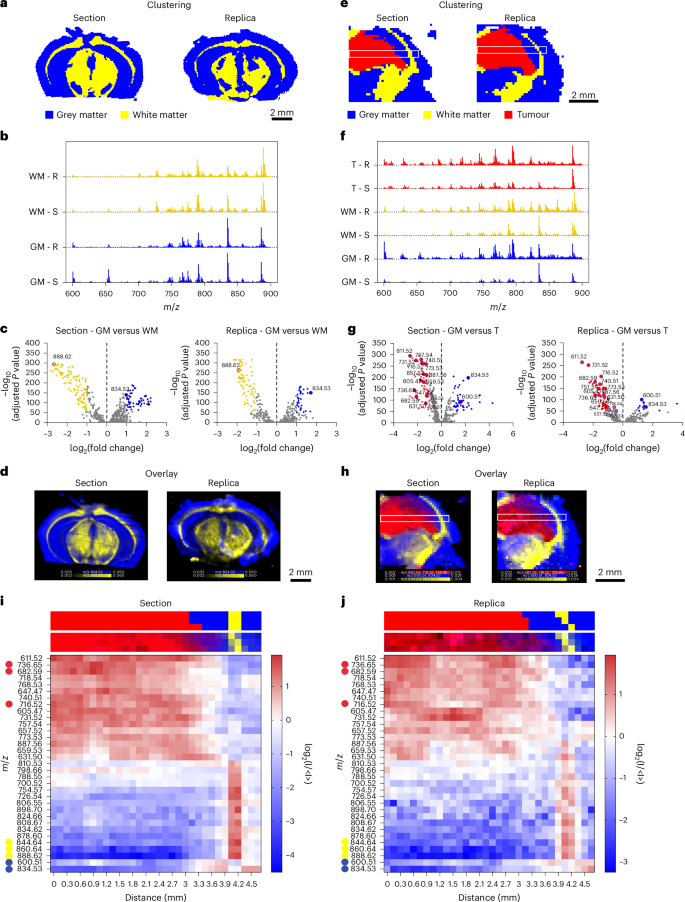
We compared the quality of lipidomic imaging on molecular replicas with its proximal tissue section5. Hierarchical cluster analysis (HCA) revealed that the top two clusters of both section and replica appeared to align with the areas of grey and white matter within the mouse brain architecture (Fig. 3a). The white matter cluster aligned with brain regions containing highly myelinated fibres, matching the expression pattern of myelin genes such as myelin basic protein in areas such as the corpus callosum. Meanwhile the grey matter cluster corresponded to regions with higher cell body densities and fewer myelinated fibres, such as the cortex4.
a, HCA maps of the top two clusters from the DESI-MSI data of a murine brain section and its molecular replica, corresponding to grey matter (blue) and white matter (yellow). b, Average spectra displaying normalized counts versus m/z, for the top two clusters associated with grey matter (GM) and white matter (WM) in section (S) and replica (R). c, Volcano plots of the differential lipid distribution between grey and white matter, highlighting the role of m/z 834.53 (PS 40:6) and 888.62 (SHexCer 42:2;O2) as grey and white matter biomarkers in both section and replica. d, An overlay map of TIC-normalized intensity for m/z 834.53 (blue) and 888.62 (yellow). e, HCA maps of the top three clusters from the DESI-MSI dataset of a tumour-bearing mouse brain section and replica, corresponding to grey matter (GM, blue), white matter (WM, yellow), and tumour (T, red). White boxes indicate regions used for lipid abundance analysis. f, Average spectra showing normalized counts versus m/z, for the three clusters associated with grey matter (GM), white matter (WM) and tumour (T) in section (S) and replica (R). g, Volcano plots comparing lipid distribution between GM and T, highlighting the most representative lipid peaks in both section and replica. h, An overlay map of cumulative TIC-normalized intensity for peaks marking grey matter (m/z 834.53 and m/z 600.51, blue), white matter (m/z 844.64, m/z 860.64 and m/z 888.62, yellow) and tumour (m/z 682.59, m/z 716.52 and m/z 736.65, red) in section and replica. The white boxes indicate regions used for lipid abundance analysis across GM, WM and T. i,j, Heatmaps showing the relative abundance of 33 lipid peaks (from analysis in g) along the major axis of the white box in e and h for the section (i) and replica (j). Top and bottom strips above each heatmap show the cluster assignment and relative lipid intensity for corresponding pixels from e and h.
The lipid composition of these two clusters confirmed this morphological classification (Fig. 3b,c). The higher abundance of m/z 888.62 (sulfated hexosylceramide (SHexCer) 42:2;O2) and lower abundance of m/z 834.53 (phosphatidylserine (PS) 40:6) in white matter were consistent with established DESI-MSI brain biomarkers26. Mapping the relative intensity of these two peaks across the sample accurately delineated white and grey matter regions in both the replica and the reference section (Fig. 3d). The congruent distribution of these lipid species across replica and section confirmed the feasibility of generating a lipidomic profile map from a nanoneedle replica of the mouse brain. Minor discrepancies in brain morphology were attributed to the inherent anatomical differences between adjacent tissue sections.
We then used the molecular replica to characterize a cancerous lesion within a mouse glioma5. The top three HCA clusters classified white matter, grey matter and tumour regions with comparable effectiveness in section and replica (Fig. 3e). When analysing a dataset including all spectra across tissue and replica, HCA similarly discriminated tumour, white matter and grey matter, highlighting the correspondence of the lipid profile on replica and section. Principal component analysis of intercluster variance further confirmed that both the replica and the section displayed similar characteristic features, enabling accurate mapping of tumour margins and transitions between tissue types (Supplementary Fig. 6).
Comparative analysis of the average mass spectra for each cluster allowed the lipid compositions of white matter, grey matter and tumour to be distinguished and the most relevant differentially abundant species to be identified (Fig. 3f,g and Supplementary Fig. 7). Again, clusters associated with white matter in both the section and replica exhibited a prominent peak at m/z 888.62 (SHexCer 42:2;O2), while the grey matter clusters showed a prominent peak at m/z 834.53 (PS 40:6). The tumour cluster showed an increase in the abundance of m/z 736.65 (ceramide (Cer) 46:1;O4), m/z 682.59 (Cer 42:0;O4) and m/z 716.52 (phosphatidylethanolamine (PE) 34:1). The intensity map for these characteristic species effectively discriminated tumour, white matter and grey matter in the replica as well as in the section (Fig. 3h), demonstrating the replica’s ability to accurately reproduce the lipidomic map of the original tissue, even in the presence of a cancerous lesion. The analysis of two additional, distinct brain samples confirmed the reproducibility of our approach (Supplementary Fig. 8).
We assessed the spatial distribution of the 33 most representative species in the section and replica by evaluating their abundance along the major axis (length) of the white box (Fig. 3h and Supplementary Fig. 9). The heatmaps from both section and replica revealed a clear and congruent pattern in lipid abundance across tumour, grey matter and white matter (Fig. 3i,j and Supplementary Fig. 9). The tumour–grey matter transition occurred at 3.1 mm from the left edge. The tumour was rich in lipids at m/z 736.65 (Cer 46:1;O4), m/z 682.59 (Cer 42:0;O4) and m/z 740.51 (phosphatidylcholine (PC) 30:0), while grey matter showed high levels of m/z 600.51 (Cer 36:1;O2) and m/z 834.53 (PS 40:6). At 4 mm, a 0.3 mm white matter layer was enriched in m/z 844.64 (PS O-41:1), m/z 860.64 (PS 41:0) and m/z 888.62 (SHexCer 42:2;O2) followed by grey matter (4.3–4.7 mm) with a lipid profile consistent with the previous grey matter region.
These data indicate that a molecular replica can map the biomolecular architecture of a tumour-bearing tissue. The quality of the replica information enables unsupervised classification of individual pixels with comparable fidelity to the original sample enabling mapping lipid abundance with comparable quality to the tissue.
Spatial lipidomics of HG replica
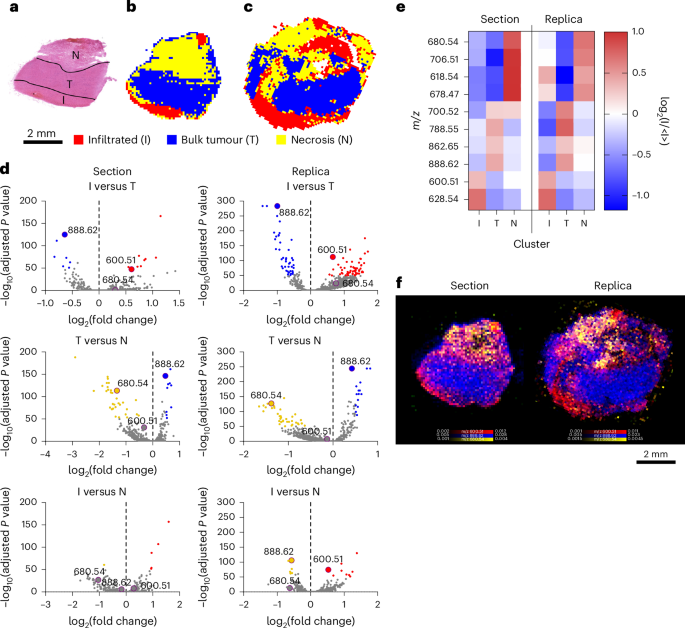
We then captured the molecular heterogeneity of a HG biopsy. Histopathological analysis identified three distinct regions of healthy brain infiltrated by tumour (I), bulk tumour (T) and necrosis (N) (Fig. 4a). HCA could distinguish these three regions and identify their lipid composition in the replica and the section (Fig. 4b–e). Mapping the distribution of the peak at m/z 600.51 (Cer 36:1;O2) characterized the infiltrated tumour, while m/z 888.62 (SHexCer 42:2;O2) distinguished the bulk tumour and m/z 680.54 (Cer 43:0;O3) preferentially associated with areas of necrosis (Fig. 4f).
a, A bright-field image of the haematoxylin and eosin (H&E)-stained section of HG biopsy, showing infiltrated tumour (I), bulk tumour (T) and necrosis (N). b,c, HCA maps of the top three clusters from DESI-MSI images of the section (b) and replica (c). d, Volcano plots showing the differential lipid abundance analysis across I, T and N. consistently identifying m/z 600.51 (Cer 36:1;O2) for I, m/z 888.62 (SHexCer 42:2;O2) for T and m/z 680.54 (Cer 43:0;O3) for N in section and replica. e, A heatmap of the ten most representative lipids defining I, T and N in both section and replica. f, An overlay map of TIC-normalized intensity for m/z 600.51 (Cer 36:1;O2, red), 888.62 (SHexCer 42:2;O2, blue) and 680.54 (Cer 43:0;O3, yellow).
Machine learning inference of HG grade
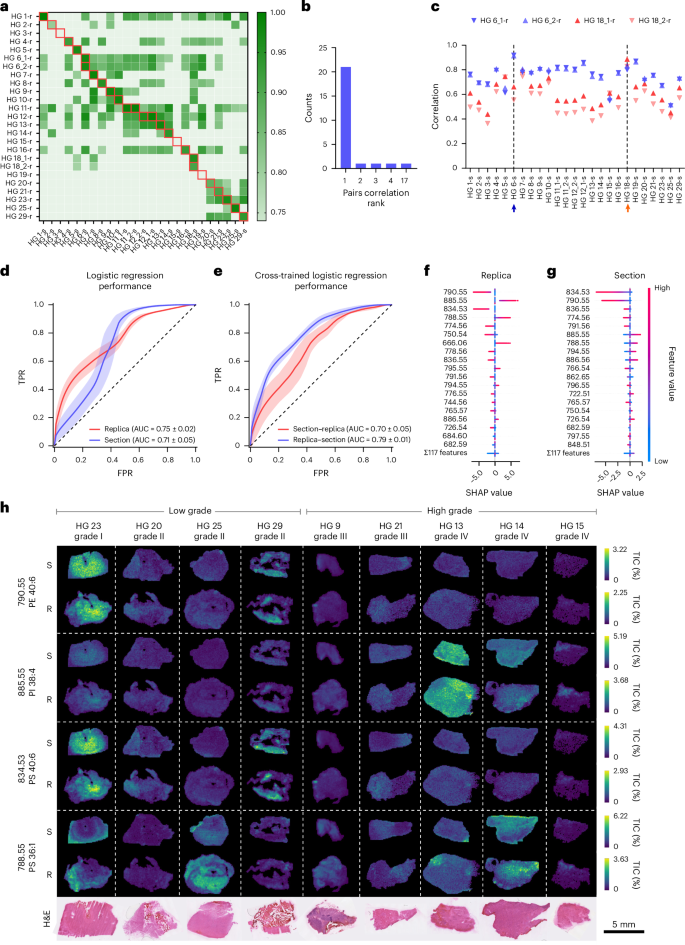
To evaluate whether replicas could match the predictive power of original tissues, we collected DESI-MSI maps of molecular replicas (HG-r) and reference sections (HG-s) from 23 HG samples with known prognoses (Supplementary Table 2). Additional sections (HG 11, 12) and replicas (HG 6, 18) were collected for intrapatient and interreplica validations, for a total of 25 sections and 25 replicas.
We observed a high correlation between replicas and their reference tissue section, indicating a preserved molecular profile (Fig. 5a). The correlation with the reference section ranked first with a probability of 0.85, and ranked in the top five with a probability of 0.96, well above the random expectations of 0.04 and 0.2, respectively (Fig. 5b). Replicas from the same section preserved a common molecular signature, showing higher correlation with their reference section than with any other (HG 6, 18) (Fig. 5c).
a, A heatmap of correlation values between 25 tissue sections and 25 replicas from 23 biopsy samples. Samples are labelled HG plus a sequential number; multiple sections or replicas from the same biopsy include a trailing underscore and number. Sections are marked with ‘s’ and replicas with ‘r’. The red boxes indicate matched section–replica pairs. b, A count plot of correlation rank between matching tissue sections and molecular replicas. c, Pearson correlation analysis of the molecular replicas HG6_1-r, HG6_2-r, HG18_1-r and HG18_2-r, with all 25 sections. The arrows mark the reference HG6-s (blue) and HG18-s (orange) section. d, Receiver operator characteristics for the logistic regression classifier applied to the HG dataset. Data are mean ± s.d. from 100 random-seed classifications. e, Receiver operator characteristics for logistic regression cross-inference: model trained on sections, tested on replicas and vice versa. Data are mean ± s.d. from 100 random-seed classifications. TPR, true positive rate; FPR, false positive rate. f,g, SHAP value distributions of top-ranked features in the replica (f) and the section model (g). Positive SHAP values indicate contribution to high-grade predictions. h, TIC-normalized ion maps of selected replicas (R) and tissue sections (S) showing the top four peaks from f, alongside H&E-stained morphology.
We next assessed disease grade prediction using sections and replicas comparing four classification models: logistic regression, decision trees, XGBoost27 and LightGBM28. All models performed better than a random model for both sections and replicas (Fig. 5d and Supplementary Fig. 10), with disease grade assignments showing a high degree of concordance. Logistic regression demonstrated superior performance (Fig. 5d and Supplementary Fig. 10) and thus was considered for further analysis. The classification performance was comparable for sections and replicas with a mean (±s.d.) area under the curve (AUC) for the receiver operating characteristic (ROC) of 0.71 ± 0.05 and 0.75 ± 0.02, respectively (Fig. 5d and Supplementary Table 3). Bootstrapping statistical validation confirmed that the observed classification performance was not due to chance (Supplementary Fig. 11). The best-performing models agreed on 25 out of the 27 section–replica pairs, further indicating that the models captured similar discriminative information from the spectral data (Supplementary Table 2). Cross-model prediction analysis applying the models trained on sections to predict the replicas achieved an AUC of 0.79 ± 0.01, while models trained on replicas to predict the sections achieved an AUC of 0.70 ± 0.05, further supporting the similarity in their molecular profiles (Fig. 5e and Supplementary Table 3). Shapley additive explanations (SHAP)29 showed that section and replica shared three of the top five most predictive peaks, while nine out of the top ten displayed the same correlation with lesion grade (Fig. 5f,g). Among the top four replica peaks, two (m/z 790.55 (PE 40:6) and m/z 834.53 (PS 40:6)) correlated with low-grade tumours and two (m/z 885.55 (phosphatidylinositol (PI) 38:4) and m/z 788.55 (PS 36:1)) with high grade. Their intensity maps across representative tumours aligned with expected tumour grades (Fig. 5h). This analysis also captured the spatial heterogeneity of tumours, identifying the lower section of HG 25—histologically infiltrated healthy brain—as low grade, enriched in the m/z values of 790.55 and 834.53, and the edges of HG 23 as high grade, enriched in m/z 788.55 and depleted in low-grade markers. These results established that the molecular replica generated using nanoneedles can replace a tissue section for disease state classification.
Molecular replicas of living tissues
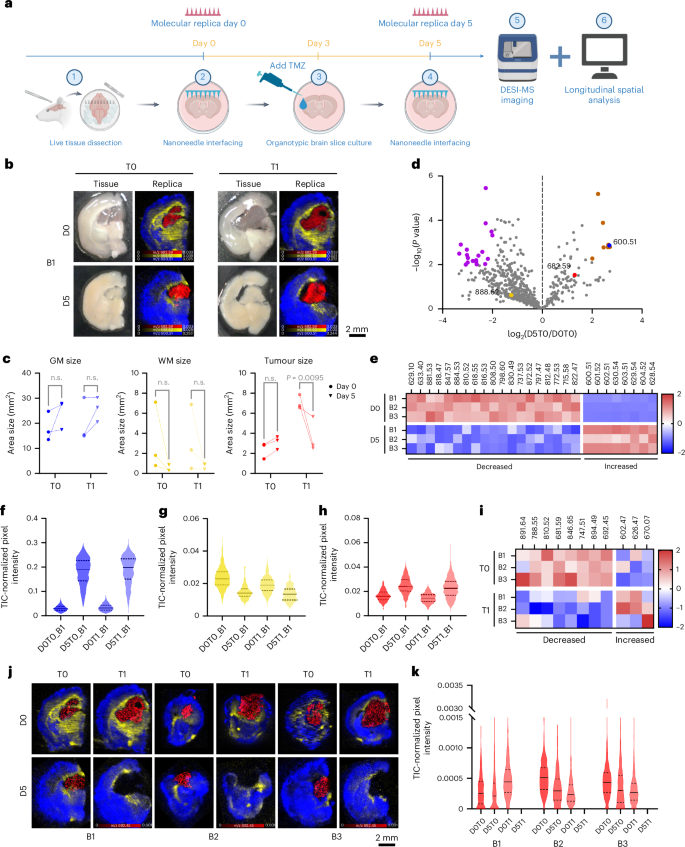
The non-destructive molecular collection enabled by nanoneedles offers a unique opportunity to sample living tissue longitudinally, allowing one to track its spatiotemporal lipidomic evolution. We tested this ability by monitoring the response of live glioma-bearing tissue slices to temozolomide (TMZ) treatment. We collected longitudinal molecular replicas on day 0 (D0T1, treated; D0T0, control) and day 5 (D5T1, treated; D5T0, control) while exposing the samples to TMZ or dimethylsulfoxide from day 3 (Fig. 6a).
a, A schematic of the longitudinal analysis workflow. Created with BioRender.com. b, Photographs (left) of brain 1 (B1) showing treated (T1) and untreated (T0) tissue slices at day 0 (D0) and day 5 (D5), with corresponding DESI-MSI maps (right) of grey matter (m/z 600.51, blue), white matter (m/z 888.62, yellow) and tumour (m/z 682.59, red) markers from molecular replicas of the photographed acute tissue slice. c, Quantification of grey matter, white matter and tumour areas from DESI-MSI molecular replicas data across time and treatment in three brain slices. Statistics: ordinary two-way ANOVA with post-hoc Šidák multiple-comparisons test. n.s., not significant. d, A volcano plot of lipid changes over time in untreated brains, highlighting markers for white matter (yellow), grey matter (blue) and tumour (red) and significantly increased (brown) or decreased (purple) species. N = 3 brain slices. e, A heatmap of the differentially abundant species (orange and purple) shown in d. f–h, Violin plots of grey matter (f), white matter (g) and tumour (h) marker abundance in brain 1 across time and treatment. Data: solid line: median; dashed lines: upper and lower quartiles. i, A heatmap of z scores for treatment-dependent significant lipids. j, DESI-MSI maps of treated and untreated molecular replicas at day 0 and day 5 for the lipid at m/z 692.45 (PS 29:0, red) in the tumour alongside grey matter (m/z 600.51, Cer 36:1;O2, blue) and white matter (m/z 888.62, SHexCer 42:2;O2, yellow) markers. k, Violin plots of m/z 692.45 (PS 29:0) abundance. Data: solid line: median; dashed lines: upper and lower quartiles.
We first assessed the feasibility of generating molecular replicas from living tissue with minimal perturbation. Nanoneedle imprinting did not impact brain slice viability immediately (Extended Data Fig. 2a,b) or over the 5 days of culture (Extended Data Fig. 2c,d), while the TMZ treatment selectively reduced the viability of cells in the tumour region by day 5 (Extended Data Fig. 2e–g). The day 0 and day 5 replicas captured the entire tissue slice area (Extended Data Fig. 2h).
Spatiotemporal lipidomics of molecular replicas
We used these longitudinal molecular replicas to study the spatiotemporal variations in the lipidomic profile in response to TMZ treatment. The DESI imaging of the molecular replicas at both timepoints showed well-defined brain regions (Fig. 6b and Supplementary Fig. 12a,c). Tumour, grey matter and white matter were distinguished by the same lipids identified for the frozen sections (Figs. 3h and 6b). The morphology of these regions aligned well with the expected structure of the tumour-bearing brain and closely matched the brain structure observed in the slices, which evolved over the 5 days in culture. Over time, and regardless of treatment, the size of the grey matter slightly increased and the white matter decreased, consistent with the expected axonal degeneration in cultured brain slices30,31 (Fig. 6c). By contrast, the tumour size slightly increased in untreated samples while decreasing significantly in response to TMZ treatment, supporting the observation that the treatment selectively targeted tumour cells (Fig. 6c and Extended Data Fig. 2g).
We then assessed the temporal evolution of lipid composition by comparing lipid abundance between untreated samples at day 0 (D0T0) and day 5 (D5T0) across the three brains. Among the 629 commonly identified lipids on day 0, 21% increased in abundance over time, while 79% decreased with 31 of them becoming undetectable (Fig. 6d). Among these, 27 lipids showed significant changes, with 8 increasing and 19 decreasing in abundance (Fig. 6e). The white matter and tumour markers did not vary significantly, while the grey matter marker abundance increased over time (Fig. 6f–h and Supplementary Fig. 12b,d). The data indicate an overall decrease in the abundance and diversity of species as a consequence of culturing the brain slices. This analysis captures brain tissue remodelling during culturing and reveals changes in lipid composition and spatial distribution within individual slices, insights made possible through longitudinal sampling enabled by nanoneedles.
We then analysed the effect of treatment specifically within the tumour. Overall, the relative abundance of 42% of lipids increased with treatment, while 58% decreased. Statistical analysis identified 11 significant lipids (Fig. 6i), with 8 decreasing and 3 increasing in abundance with TMZ treatment. The most significant, m/z 692.45 (PS 29:0), selectively disappeared in treated tumours across all samples (Fig. 6j,k). Other lipid species showed distinct, treatment-dependent abundance patterns, highlighting the power of longitudinal molecular replica analysis to detect lipid-specific and specimen-specific treatment responses (Supplementary Fig. 13). Phosphatidylserines (PS29:0, PS 42:8, PS O-41:0, PS 38:4, PS 36:1) and phosphatidylinositol (PI O-39:1) featured prominently among the species decreasing in response to TMZ (Supplementary Table 1). Phosphatidylserine upregulation is a central feature of glioblastoma multiforme32 that is modulated by TMZ treatment33, including through ferroptosis34,35,36. The PI3K–AKT is a ubiquitously upregulated pathway in glioblastoma37, which is leveraged by TMZ38.
These data show that TMZ treatment alters tumour lipid composition and distribution, notably by selectively reducing specific metabolites. Using longitudinal data from molecular replicas, we characterized the response of the tumour and identified target lipids previously linked to glioma and TMZ. This analysis offers valuable insights into metabolic pathways affected by the treatment and potential biomarkers of therapeutic efficacy.